suppressPackageStartupMessages({
library(tidyverse)
library(tidymodels)
library(survival)
library(cowplot)
library(future)
library(furrr)
library(censored)
library(shapviz)
library(fastshap)
})
theme_set(theme_cowplot())
options(repr.plot.width = 15, repr.plot.height = 9)
set.seed(42)
# the multi model fitting process will use multiple cores
plan(multicore, workers = 6)
Survival prediction models, part 2#
In part 1 we learned how to train different survival models and compare them with the concordance index.
However there are other survival metrics that we might want to consider, depending on the context, in this notebook we will explore these metrics.
Dataset preprocessing#
This time we will use the censored::time_to_million dataset:
head(time_to_million)
| title | time | event | released | released_theaters | distributor | year | rated | runtime | action | ⋯ | mandarin | usa | india | mexico | uk | france | china | canada | japan | australia |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <date> | <dbl> | <fct> | <dbl> | <fct> | <dbl> | <dbl> | ⋯ | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| 10 Cloverfield Lane | 0.111342778318533 | 1 | 2016-03-11 | 3427 | paramount_pi | 2016 | pg_13 | 103 | 1 | ⋯ | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 102 Not Out | 10.654971015124438 | 1 | 2018-05-04 | 102 | sony_pictures | 2018 | pg | 102 | 0 | ⋯ | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 Strong | 0.176278921201207 | 1 | 2018-01-19 | 3018 | warner_bros | 2018 | r | 130 | 1 | ⋯ | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 idiotas | 9.406626540687972 | 1 | 2017-06-02 | 349 | lionsgate | 2017 | pg_13 | 106 | 0 | ⋯ | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 47 Meters Down | 0.228973007974901 | 1 | 2017-06-16 | 2471 | entertainmen | 2017 | pg_13 | 89 | 0 | ⋯ | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 7 Days in Entebbe | 1.713170877091160 | 1 | 2018-03-16 | 838 | focus_features | 2018 | pg_13 | 107 | 1 | ⋯ | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
data <-
# remove rows with missing data
na.omit(time_to_million) |>
# the surv variable will be our outcome
mutate(surv=Surv(time, event))
dim(data)
- 551
- 50
# separate 20% of the dataset for testing the models later
data_split <- initial_split(data, prop = 0.8, strata=time)
# create a recipe, to prepare the data for fitting
rec <- recipe(surv ~ ., data=data) |>
# remove uneeded variables
step_rm(time, event, title, released) |>
# convert factors to integers
step_integer(all_nominal_predictors()) |>
# remove variables with zero variance
step_zv(all_predictors()) |>
# center and scale all numeric variables
step_normalize(all_numeric_predictors())
Model definitions#
models <- list(
cox_ph_survival = proportional_hazards(engine='survival'),
cox_ph_glmnet = proportional_hazards(penalty=tune(), mixture=tune(), engine='glmnet'),
survreg_flexsurv = survival_reg(engine='flexsurv'),
rand_forest_partykit = rand_forest(trees = tune(), engine='partykit'),
rand_forest_aorsf = rand_forest(trees = tune(), engine='aorsf'),
decision_tree_partykit = decision_tree(engine='partykit'),
boost_tree_mboost = boost_tree(trees = tune(), engine='mboost')
) |>
map(~set_mode(.x,'censored regression'))
wsets <- workflow_set(
preproc=list(rec),
models=models
)
Model fitting#
# 5-fold cross-validation resampling for the tunning and comparisons
folds <- vfold_cv(training(data_split), v=5, strata=time)
Some metrics are only evaluated at specific time points, for example,
you might be interested in a model that can predict 1-year survival of cancer patients undergoing a treatment,
so its always important to consider the question you want to answer.
Here we will partition the time points with quantiles, but we could have used equally spaced time points instead, depends on your data.
eval_time_points <-
with(training(data_split),
quantile(time, p=seq(0.01, 0.99, length.out = 20))
)
eval_time_points
- 1%
- 0.0151560308243856
- 6.157895%
- 0.0424028085643098
- 11.31579%
- 0.0654215224706485
- 16.47368%
- 0.0910562799996761
- 21.63158%
- 0.10910910525672
- 26.78947%
- 0.140891877015565
- 31.94737%
- 0.172892207960269
- 37.10526%
- 0.200354610941643
- 42.26316%
- 0.233666944396109
- 47.42105%
- 0.264861003383604
- 52.57895%
- 0.333096401335164
- 57.73684%
- 0.429883980992403
- 62.89474%
- 0.597292977256363
- 68.05263%
- 0.738947868445357
- 73.21053%
- 1.14859426585908
- 78.36842%
- 1.72592274879734
- 83.52632%
- 2.62600273701295
- 88.68421%
- 7.18309059689147
- 93.84211%
- 10.7500402599578
- 99%
- 42
res <- workflow_map(
wsets,
"tune_grid",
seed = 42,
grid = 5,
resamples = folds,
# these metrics will be calculated for each fit
metrics=metric_set(brier_survival_integrated, brier_survival, roc_auc_survival, concordance_survival),
# some metrics are evaluated at specific time points
eval_time = eval_time_points
)
tune_metrics <- collect_metrics(res)
This time we added more metrics to evaluate, each metric is calculated differently, its important to understand how each of these metrics work and what you want to predict in order to choose the best metric for your data:
roc_auc: measures the area under the ROC curve for each of the chosen time points
brier_score: mean squared error for each time points, lower is better.
concordance index: measures if the time predictions are concordant (events are ordered correctly)
integrated brier: summary of the brier score over all given time points, lower is better.
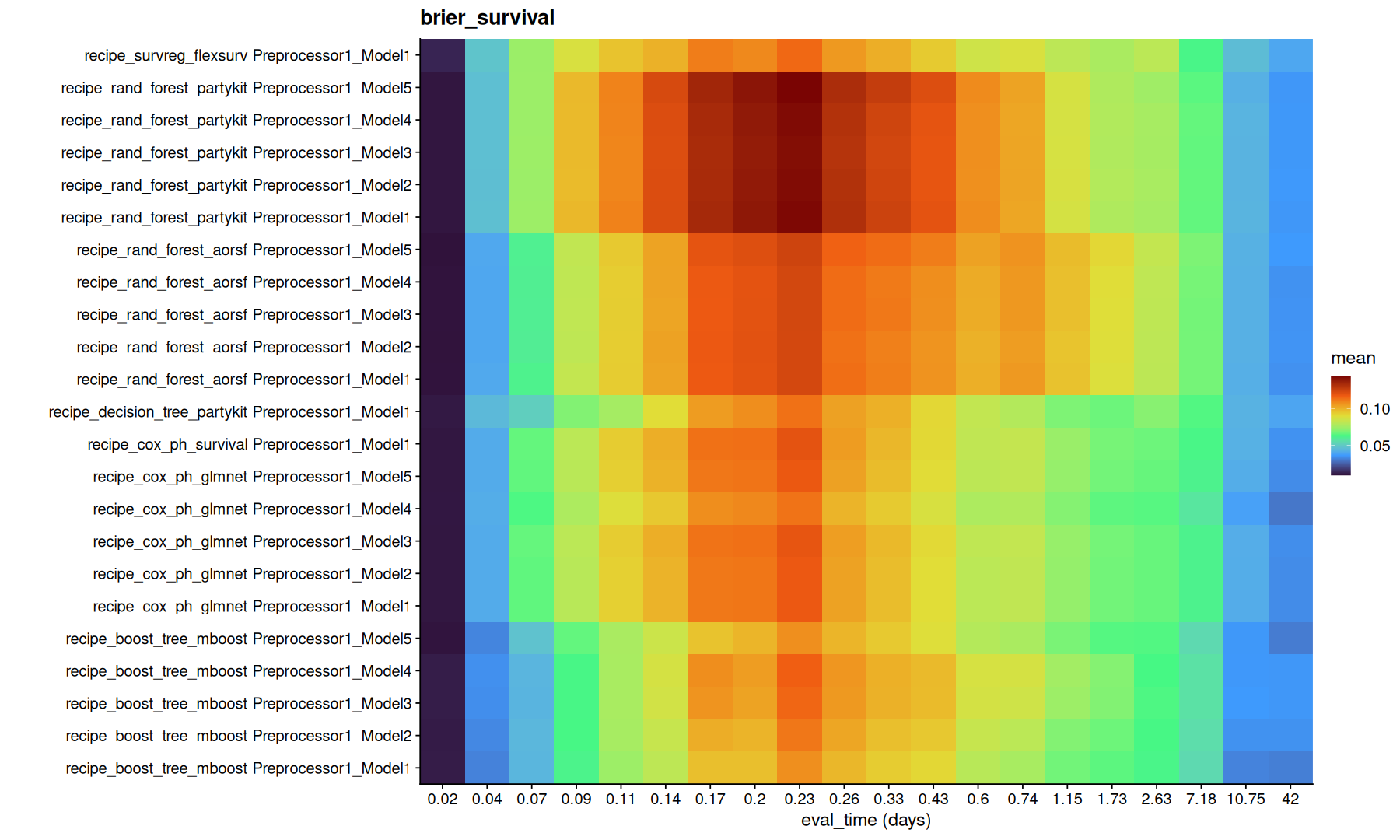
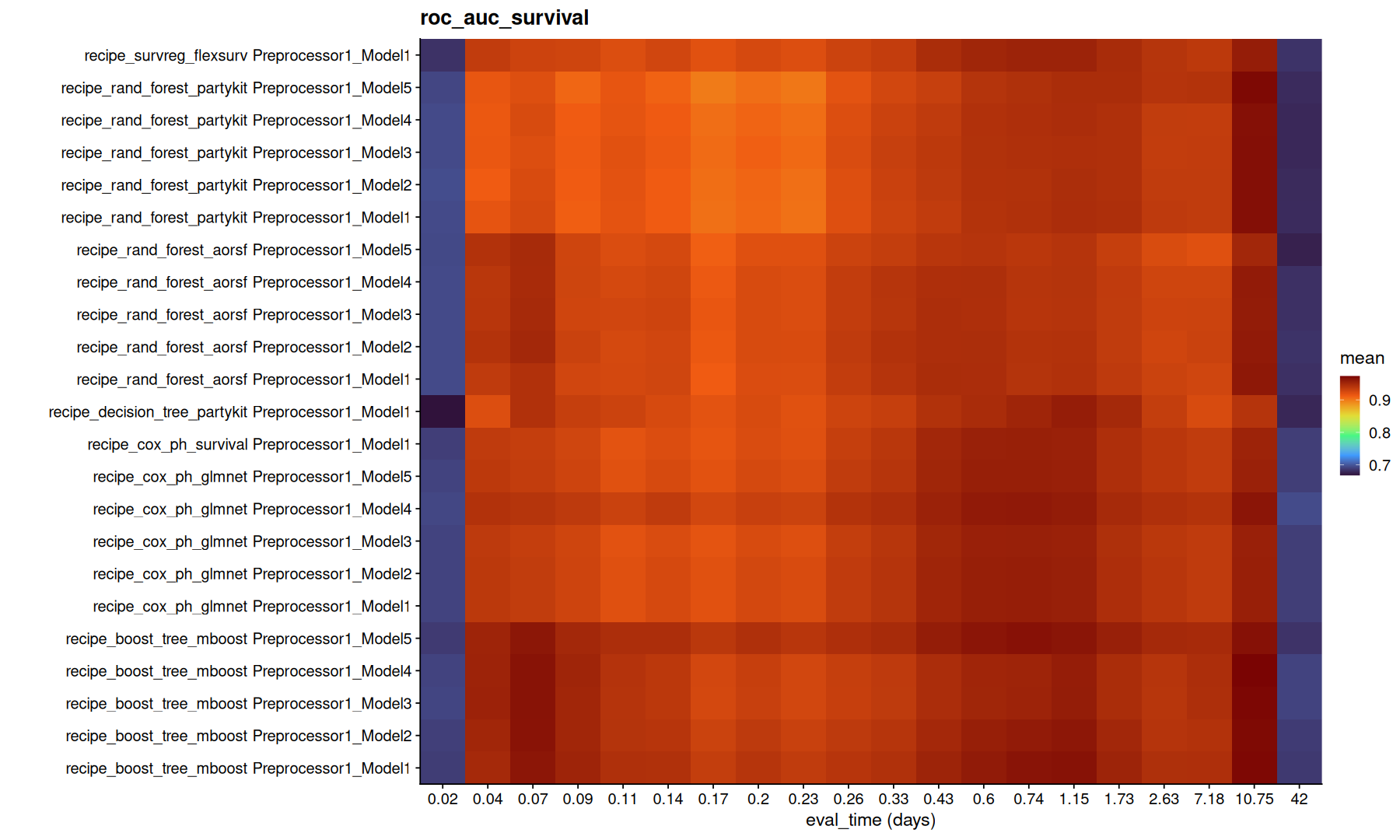
tune_metrics |>
filter(!is.na(.eval_time)) |>
group_by(.metric) |>
group_map(function(df, g) {
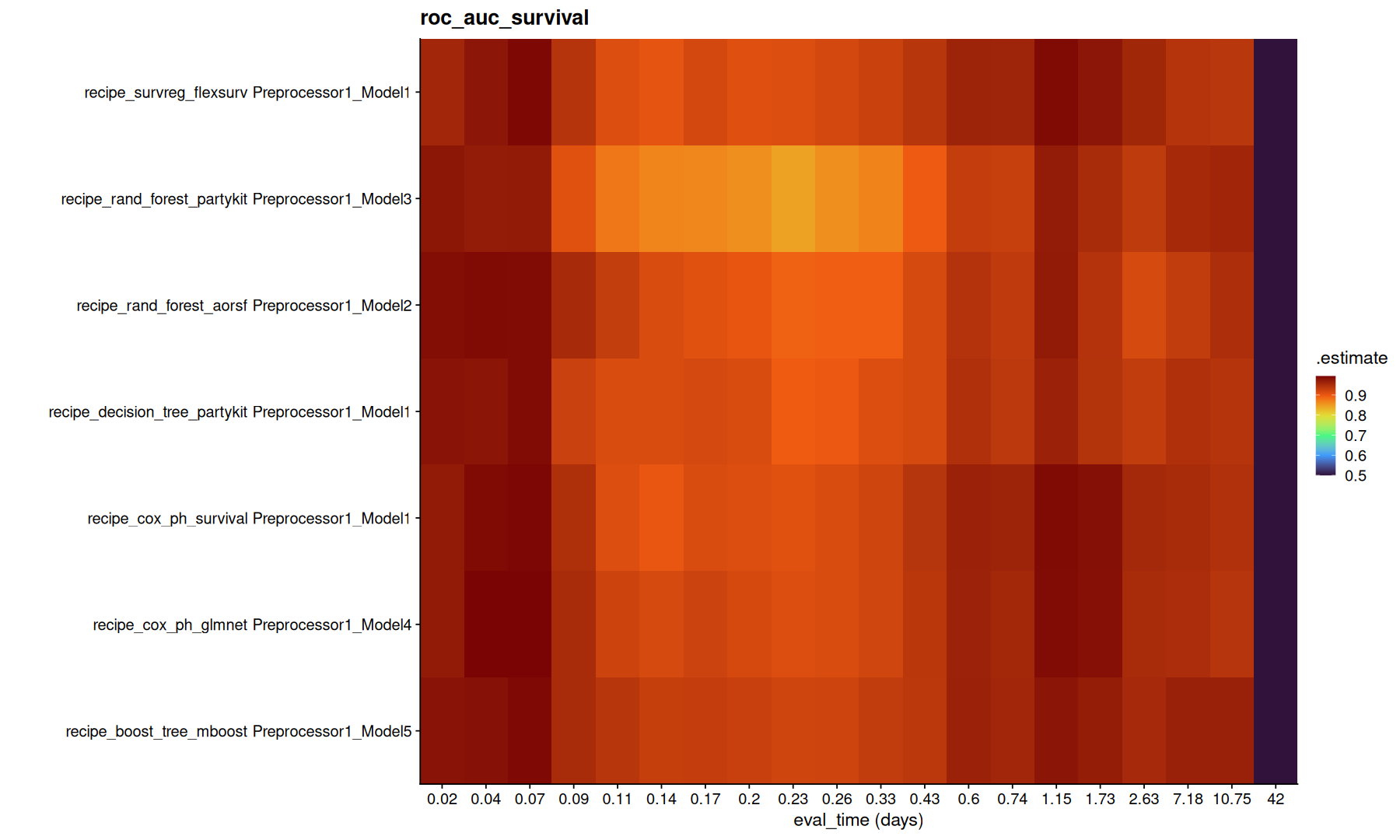
ggplot(df,aes(x=factor(round(.eval_time,2)), y=paste(wflow_id, .config), fill=mean)) +
geom_tile() +
coord_cartesian(expand=0) +
scale_fill_viridis_c(option='turbo') + labs(title=g, x='eval_time (days)', y='')
})
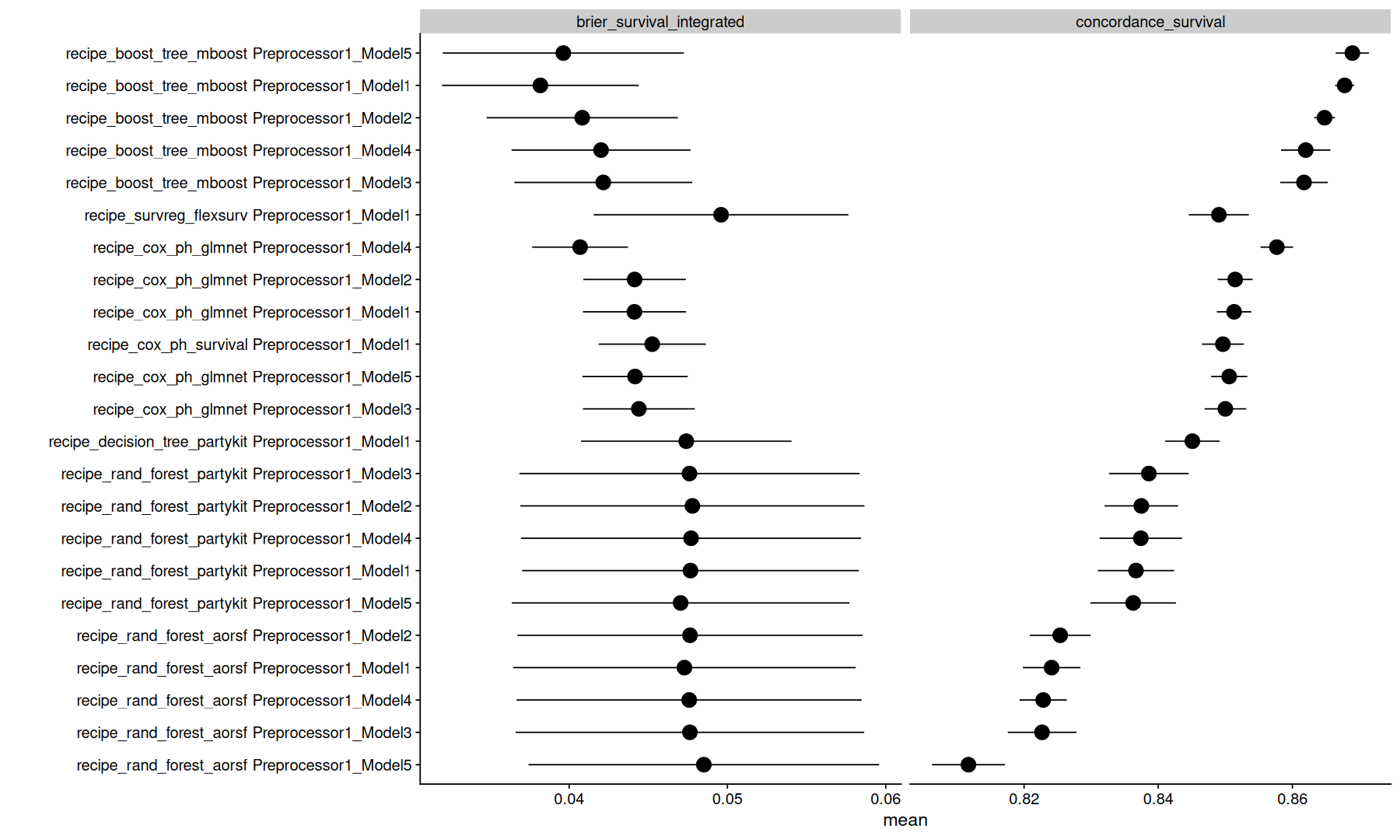
tune_metrics |>
filter(is.na(.eval_time)) |>
arrange(.metric, mean) |>
ggplot(aes(y=fct_reorder(paste(wflow_id, .config), mean), x=mean)) +
geom_point(size=5) +
geom_pointrange(aes(xmin=mean-std_err, xmax=mean+std_err)) +
facet_wrap(~.metric, scales='free_x') +
labs(y='')
Note that, the model with lowest integrated brier might not necessarly be the same as the model with highest concordance.
Again, its important to understand the difference between these metrics to decide which metric is better to optimize.
Fit the best models with all the training data#
final_fits <-
# map across each model in the table above
set_names(unique(tune_metrics$wflow_id)) |>
future_map(function(wi) {
# select the best model config, in case of tunnable parameters
bwr <- extract_workflow_set_result(res, wi)
best_params <- select_best(bwr, metric='concordance_survival')
bw <- finalize_workflow(
extract_workflow(res, wi),
best_params
)
# re-fit the best config with the initial 80% train / 20% test
fit <- last_fit(
bw,
split = data_split,
metrics = metric_set(brier_survival_integrated, brier_survival, roc_auc_survival, concordance_survival),
eval_time = eval_time_points
)
metrics <- collect_metrics(fit) |>
mutate(wflow_id = wi, .config = best_params$.config)
list(fit=list(fit), metrics=metrics)
}, .options = furrr_options(seed = T)) |>
list_transpose()
list_rbind(final_fits$metrics) |>
inner_join(tune_metrics) |>
filter(is.na(.eval_time)) |>
ggplot(aes(y=fct_reorder(wflow_id,mean), x=mean)) +
geom_point(size=4) +
geom_point(aes(x=.estimate), shape=3, size=3, stroke=3, color='darkred') +
geom_pointrange(aes(xmin=mean-std_err, xmax=mean+std_err)) +
labs(x='estimate', y='models') +
facet_wrap(~.metric, scales='free_x')
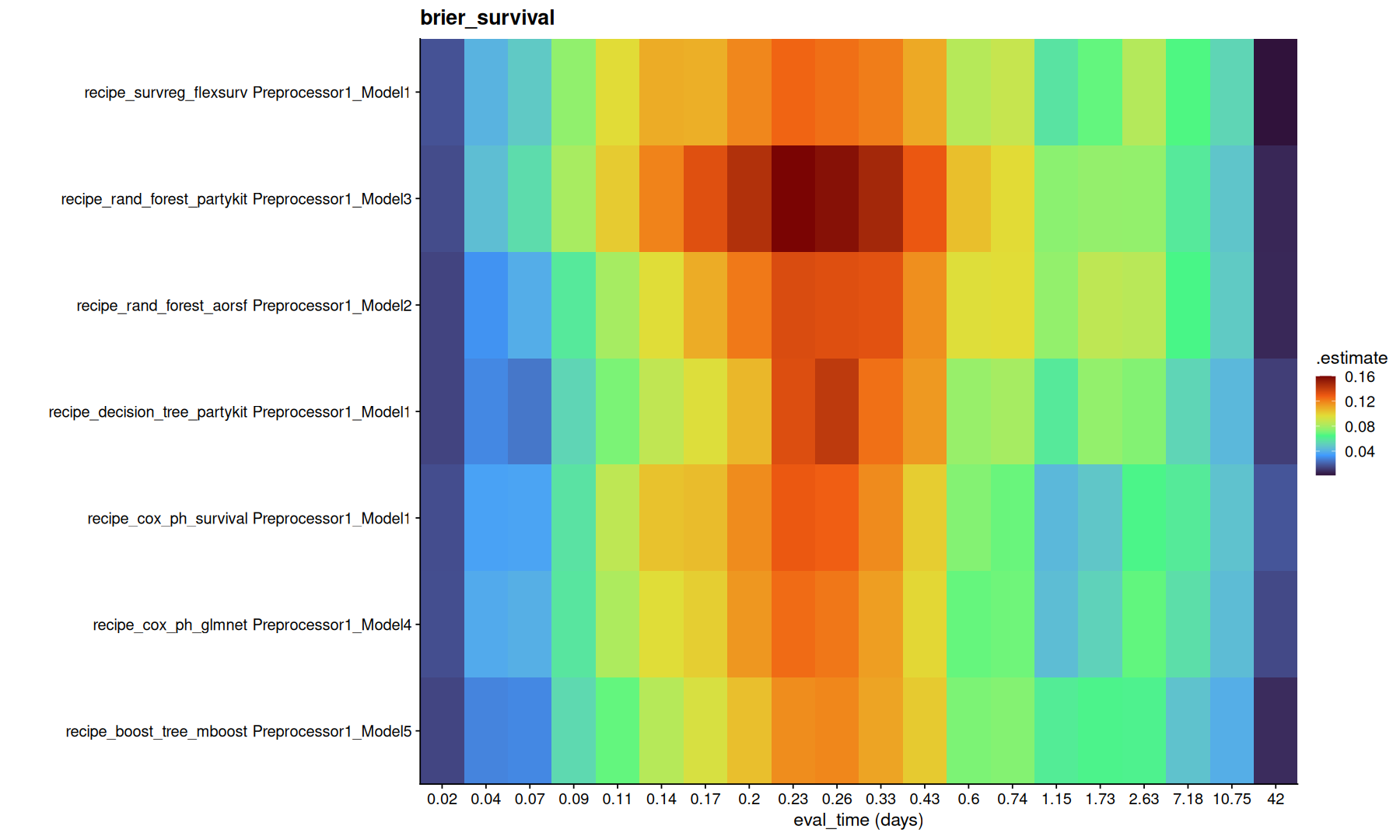
list_rbind(final_fits$metrics) |>
inner_join(tune_metrics) |>
filter(!is.na(.eval_time)) |>
group_by(.metric) |>
group_map(function(df, g) {
ggplot(df,aes(x=factor(round(.eval_time,2)), y=paste(wflow_id, .config), fill=.estimate)) +
geom_tile() +
coord_cartesian(expand=0) +
scale_fill_viridis_c(option='turbo') + labs(title=g, x='eval_time (days)', y='')
})
Variable Importance#
names(final_fits$fit)
- 'recipe_cox_ph_survival'
- 'recipe_cox_ph_glmnet'
- 'recipe_survreg_flexsurv'
- 'recipe_rand_forest_partykit'
- 'recipe_rand_forest_aorsf'
- 'recipe_decision_tree_partykit'
- 'recipe_boost_tree_mboost'
final_fits$fit[['recipe_cox_ph_glmnet']] |>
extract_spec_parsnip()
Proportional Hazards Model Specification (censored regression)
Main Arguments:
penalty = 0.0238338401768485
mixture = 0.843754897918552
Computational engine: glmnet
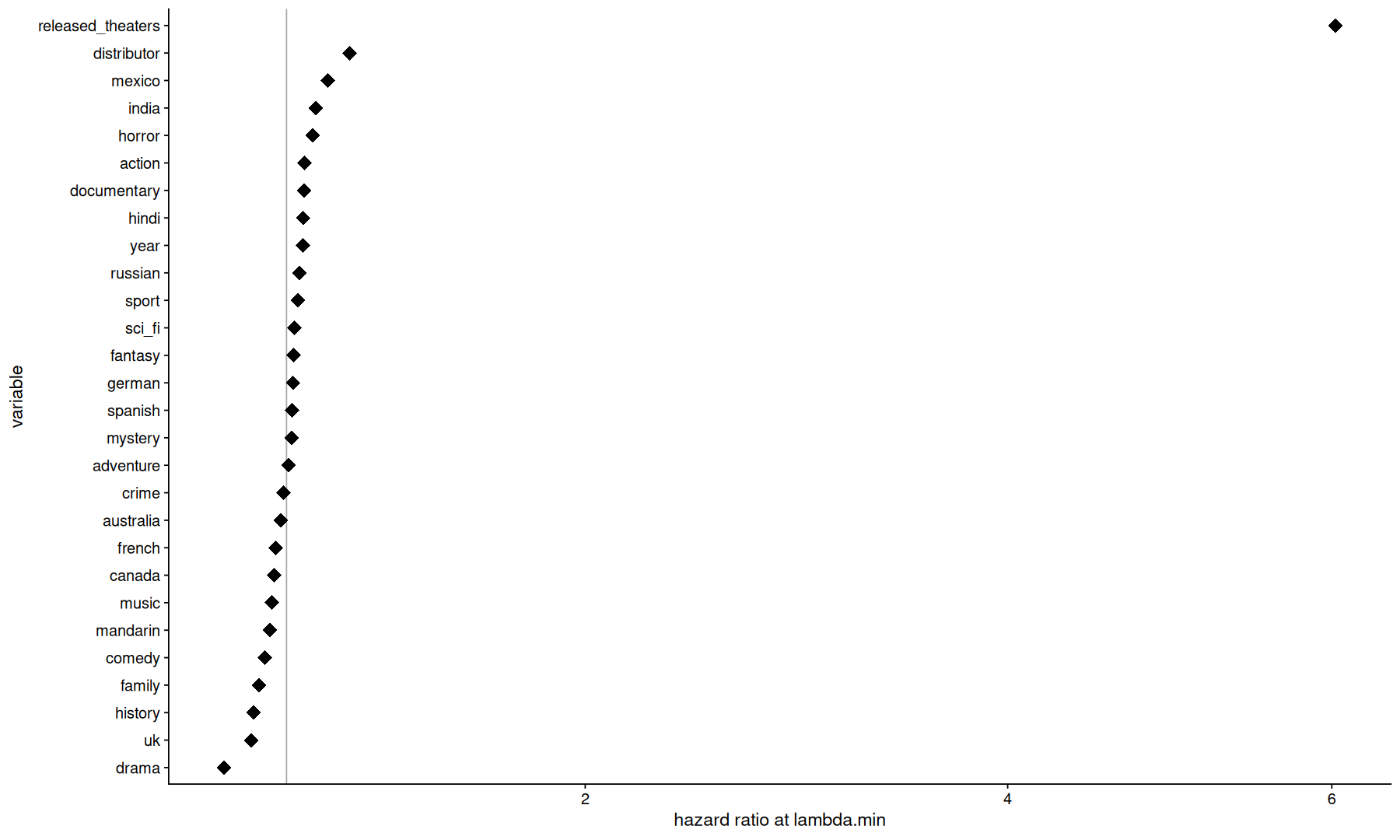
final_fits$fit[['recipe_cox_ph_glmnet']] |>
extract_fit_engine() |>
tidy() |>
filter(abs(lambda - 0.0238338401768485)<0.001) |>
mutate(exp.estimate=exp(estimate)) |>
ggplot(aes(x=exp.estimate, y=fct_reorder(term, estimate))) +
geom_vline(xintercept=1, color='darkgray') +
geom_point(size=5, shape=18) +
scale_x_sqrt() +
labs(x='hazard ratio at lambda.min', y='variable')
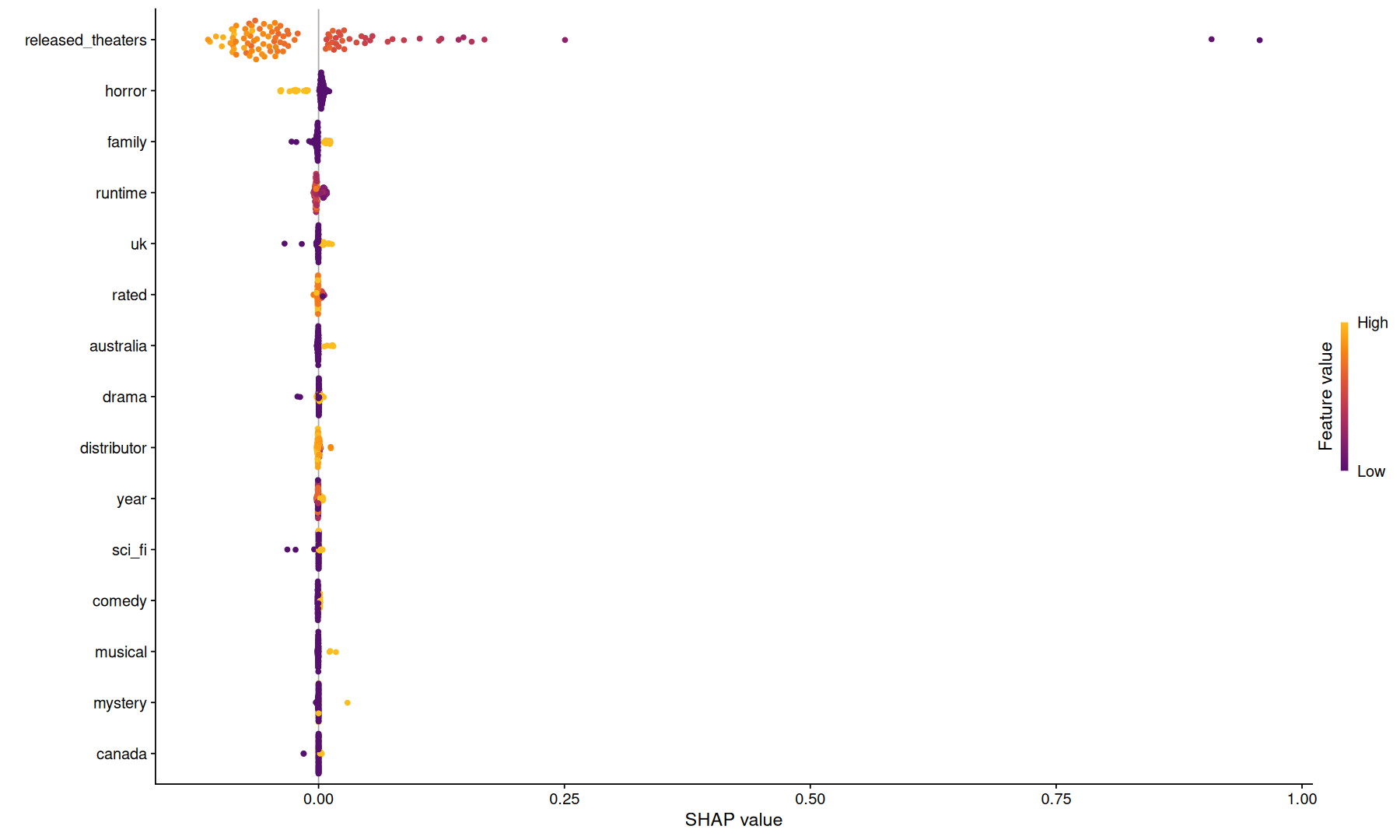
final_fits$fit[['recipe_boost_tree_mboost']] |>
extract_spec_parsnip()
Boosted Tree Model Specification (censored regression)
Main Arguments:
trees = 94
Computational engine: mboost
mboost_obj <-
final_fits$fit[['recipe_boost_tree_mboost']] |>
extract_fit_parsnip()
prec <-
final_fits$fit[['recipe_boost_tree_mboost']] |>
extract_recipe()
train_data <-
bake(prec,new_data=training(data_split), all_predictors())
library(doFuture)
registerDoFuture()
shap.vals <- fastshap::explain(mboost_obj, X = head(train_data,100), nsim=100, pred_wrapper = function(obj,newdata) pull(predict(obj,new_data=newdata),.pred_time), shap_only=FALSE, parallel=TRUE)
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-1') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-2') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-3') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-4') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-5') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."
Warning message:
"UNRELIABLE VALUE: One of the foreach() iterations ('doFuture-6') unexpectedly generated random numbers without declaring so. There is a risk that those random numbers are not statistically sound and the overall results might be invalid. To fix this, use '%dorng%' from the 'doRNG' package instead of '%dopar%'. This ensures that proper, parallel-safe random numbers are produced via the L'Ecuyer-CMRG method. To disable this check, set option 'doFuture.rng.onMisuse' to "ignore"."