suppressPackageStartupMessages({
library(tidyverse)
library(cowplot)
library(aplot)
library(ggtree)
})
theme_set(theme_cowplot())
options(repr.plot.width=15,repr.plot.height=9)
Clustered Heatmap#
There are many packages that generate clustered heatmaps on ggplot2, but none of them are composable.
This notebook illustrates how to make a clustered heatmap in a composable way, using the aplot package.
code#
insert_generic <- function(data, plot, side, size=1) {
if(side == 'top') {
insert_top(data, plot, height=size)
} else if(side == 'bottom') {
insert_bottom(data, plot, height=size)
} else if(side == 'left') {
insert_left(data, plot, width=size)
} else if(side == 'right') {
insert_right(data, plot, width=size)
} else {
stop('unknown side')
}
}
insert_hclust <- function(ggp,
cluster_by,
side=c('top','left'),
dist_method="euclidean",
hclust_method='complete',
size=0.1) {
plot <- ggp
if('aplot' %in% class(ggp)) {
ggp <- plot$plotlist[[1]]
}
mdata <-
ggp$data |>
mutate(!!!ggp$mapping) |>
pivot_wider(names_from = 'x', values_from = {{cluster_by}}, id_cols=y) |>
column_to_rownames('y') |>
as.matrix()
reduce(side, function(p, s) {
if(s %in% c('left','right')) {
dd <-
mdata |>
dist(method=dist_method) |>
hclust(method=hclust_method) |>
ggtree(branch.length='none')
if(s == 'right') { dd <- dd + scale_x_reverse() }
p <- insert_generic(p, dd, side=s, size=size)
}
if(s %in% c('top','bottom')) {
dd <-
t(mdata) |>
dist(method=dist_method) |>
hclust(method=hclust_method) |>
ggtree(branch.length='none')
if(s == 'bottom') { dd <- dd + coord_flip() }
else { dd <- dd + layout_dendrogram() }
p <- insert_generic(p, dd, side=s, size=size)
}
return(p)
}, .init=plot)
}
insert_annot <- function(ggp, side, var, size=0.02, ggextra=NULL) {
plot <- ggp
if('aplot' %in% class(ggp)) {
ggp <- plot$plotlist[[1]]
}
rem_axis <- list(coord_cartesian(expand=0) , theme(
line=element_blank(),
rect=element_blank(),
axis.text=element_blank(),
axis.title = element_blank(),
plot.margin = margin(t = 0, r = 0,b = 0,l = 0)
))
if(side %in% c('top','bottom')) {
p <-
ggplot(ggp$data, aes(x=!!ggp$mapping$x, y=1, fill={{var}})) +
geom_tile() + rem_axis + ggextra
}
if(side %in% c('left','right')) {
p <-
ggplot(ggp$data, aes(y=!!ggp$mapping$y, x=1, fill={{var}})) +
geom_tile() + rem_axis + ggextra
}
insert_generic(data=plot, plot=p, side=side, size=size)
}
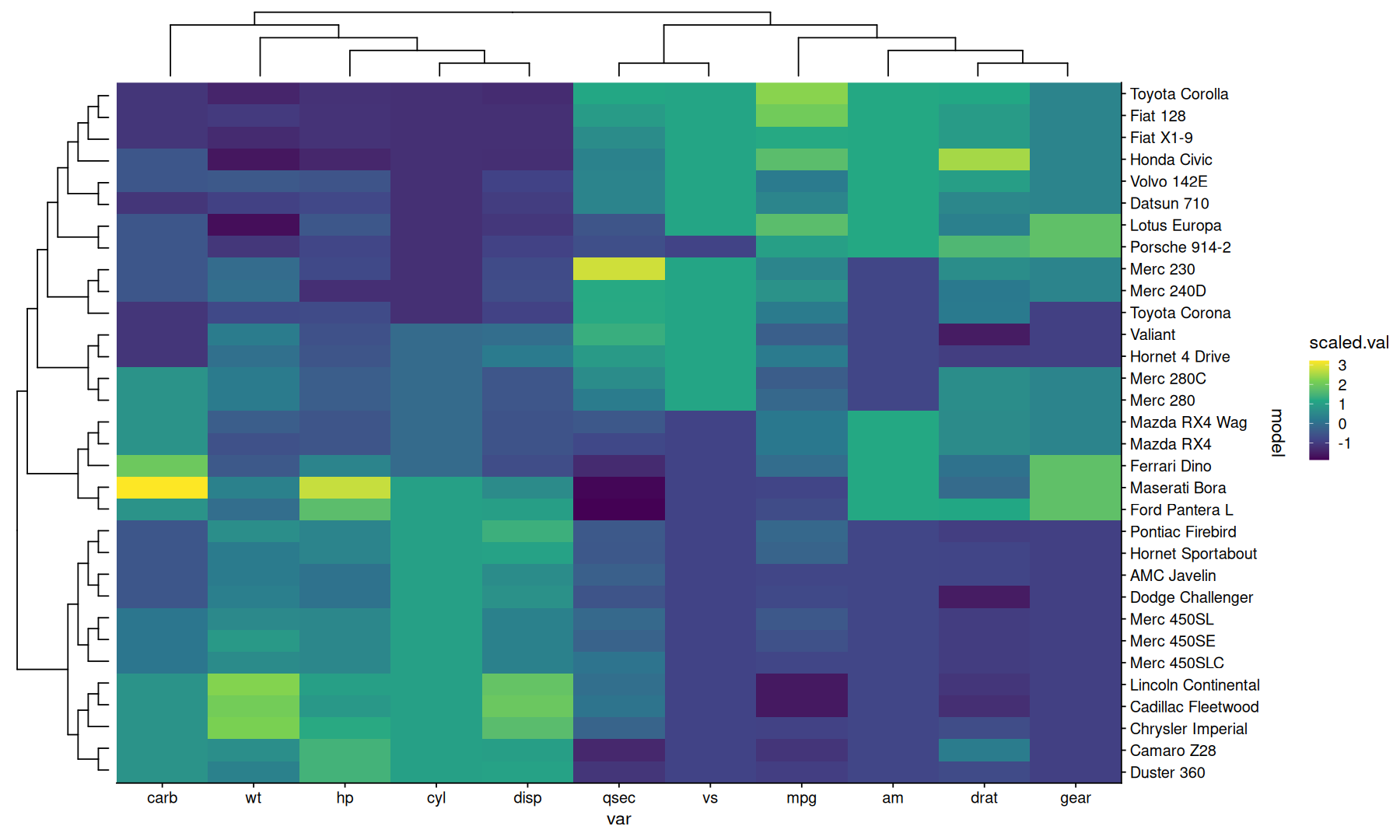
Basic Heatmap#
data <-
rownames_to_column(mtcars, 'model') |>
pivot_longer(names_to = 'var', values_to = 'value', -model) |>
mutate(scaled.val = scale(value), .by=var)
head(data, 3)
| model | var | value | scaled.val |
|---|---|---|---|
| <chr> | <chr> | <dbl> | <dbl[,1]> |
| Mazda RX4 | mpg | 21 | 0.150884824647657 |
| Mazda RX4 | cyl | 6 | -0.104987808575239 |
| Mazda RX4 | disp | 160 | -0.570619818667904 |
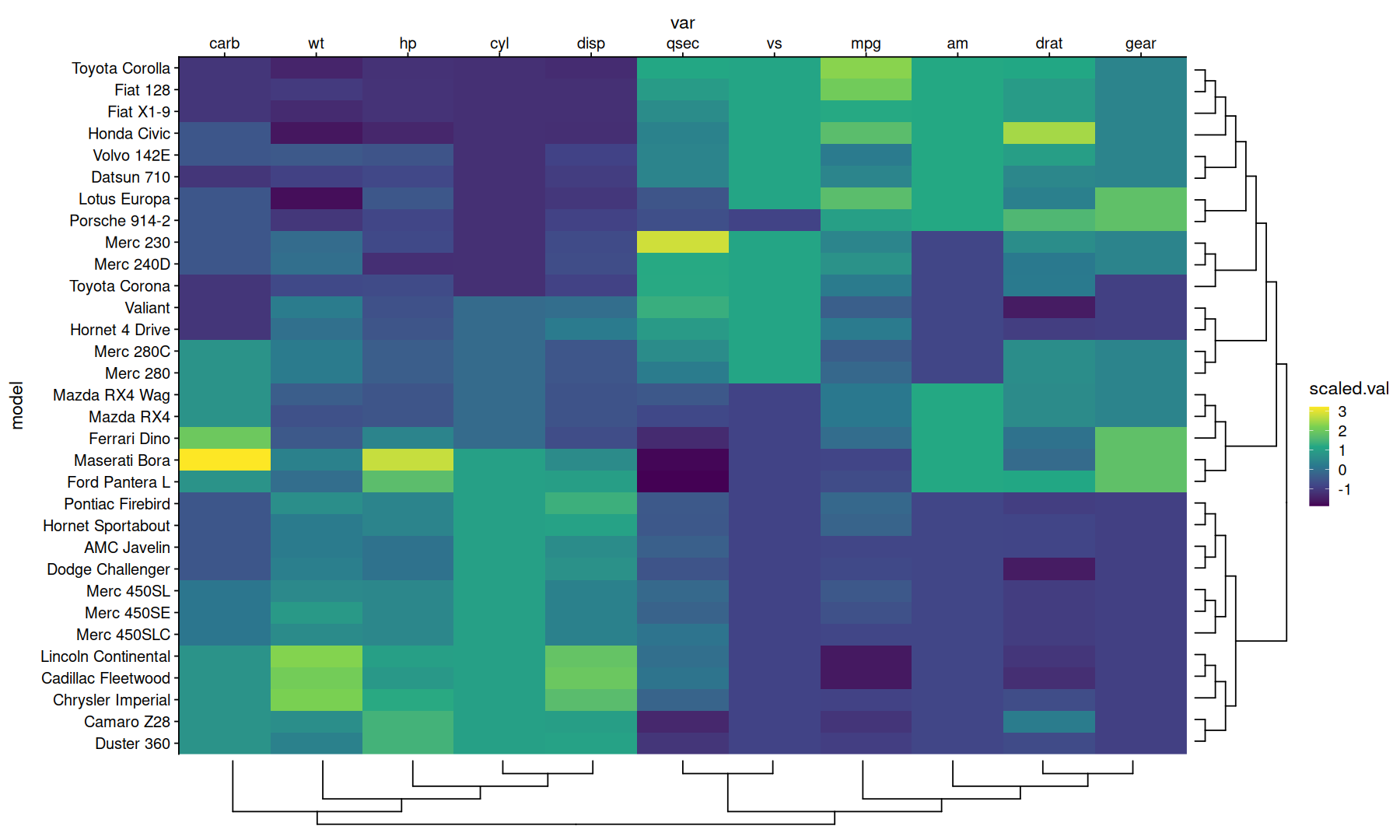
Dendrogram on the alternate side#
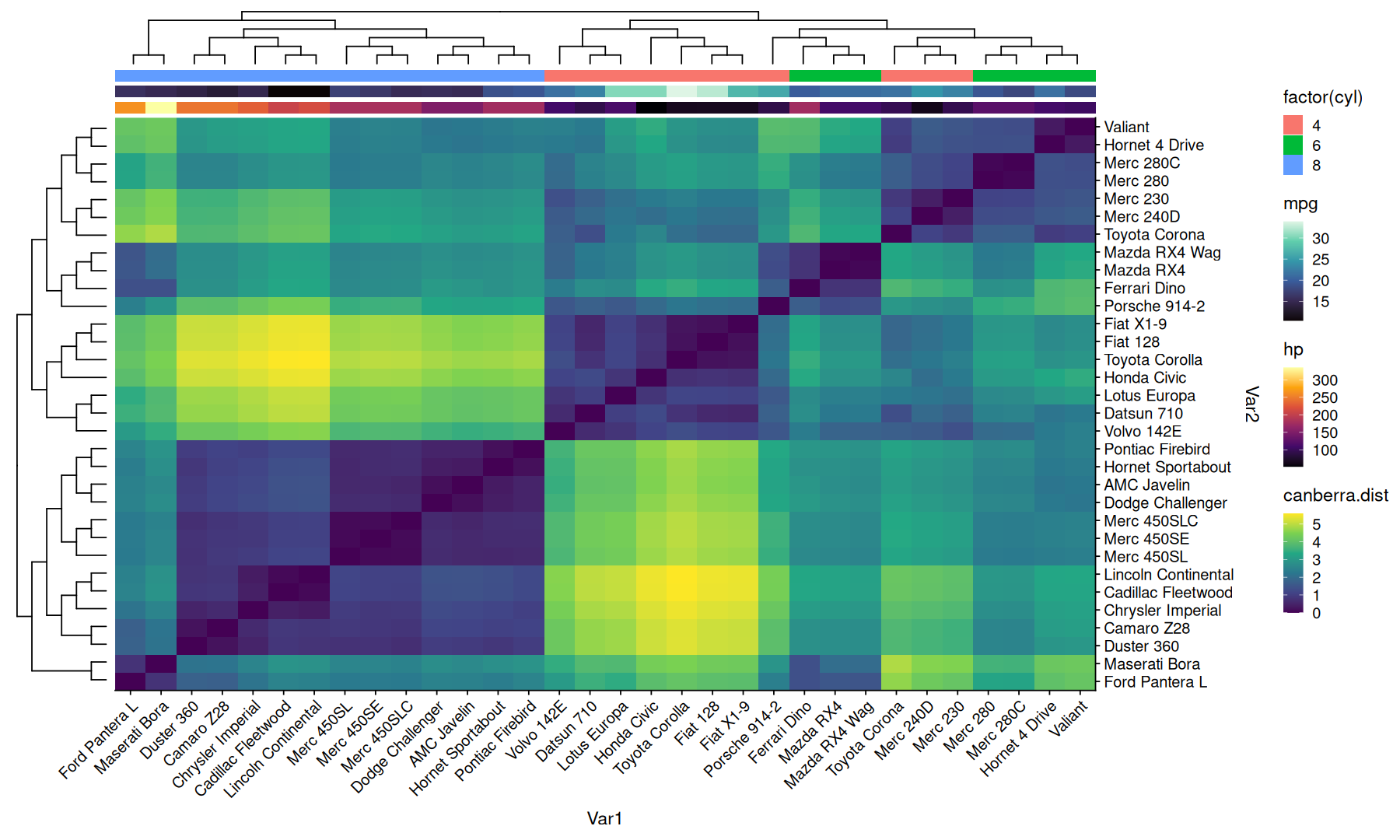
Heatmap with annotations#
data <-
dist(mtcars, method='canberra') |>
as.matrix() |>
as.data.frame.table(responseName='canberra.dist') |>
inner_join(rownames_to_column(mtcars,'model'), by=c('Var1'='model'))
head(data, 3)
| Var1 | Var2 | canberra.dist | mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <fct> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | Mazda RX4 | Mazda RX4 | 0.0000000000000000 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 2 | Mazda RX4 Wag | Mazda RX4 | 0.0694454500289716 | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 3 | Datsun 710 | Mazda RX4 | 2.2473559008738091 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
(
data |>
ggplot(aes(x=Var1, y=Var2, fill=canberra.dist)) +
geom_tile() +
coord_cartesian(expand=0) +
scale_y_discrete(position='right') +
theme(axis.text.x=element_text(angle=45,vjust=1,hjust=1)) +
scale_fill_viridis_c()
) |>
insert_annot(side = 'top', var = hp, ggextra=scale_fill_viridis_c(option='inferno')) |>
insert_annot(side = 'top', var = mpg, ggextra=scale_fill_viridis_c(option='mako')) |>
insert_annot(side = 'top', var = factor(cyl)) |>
insert_hclust(cluster_by=canberra.dist)
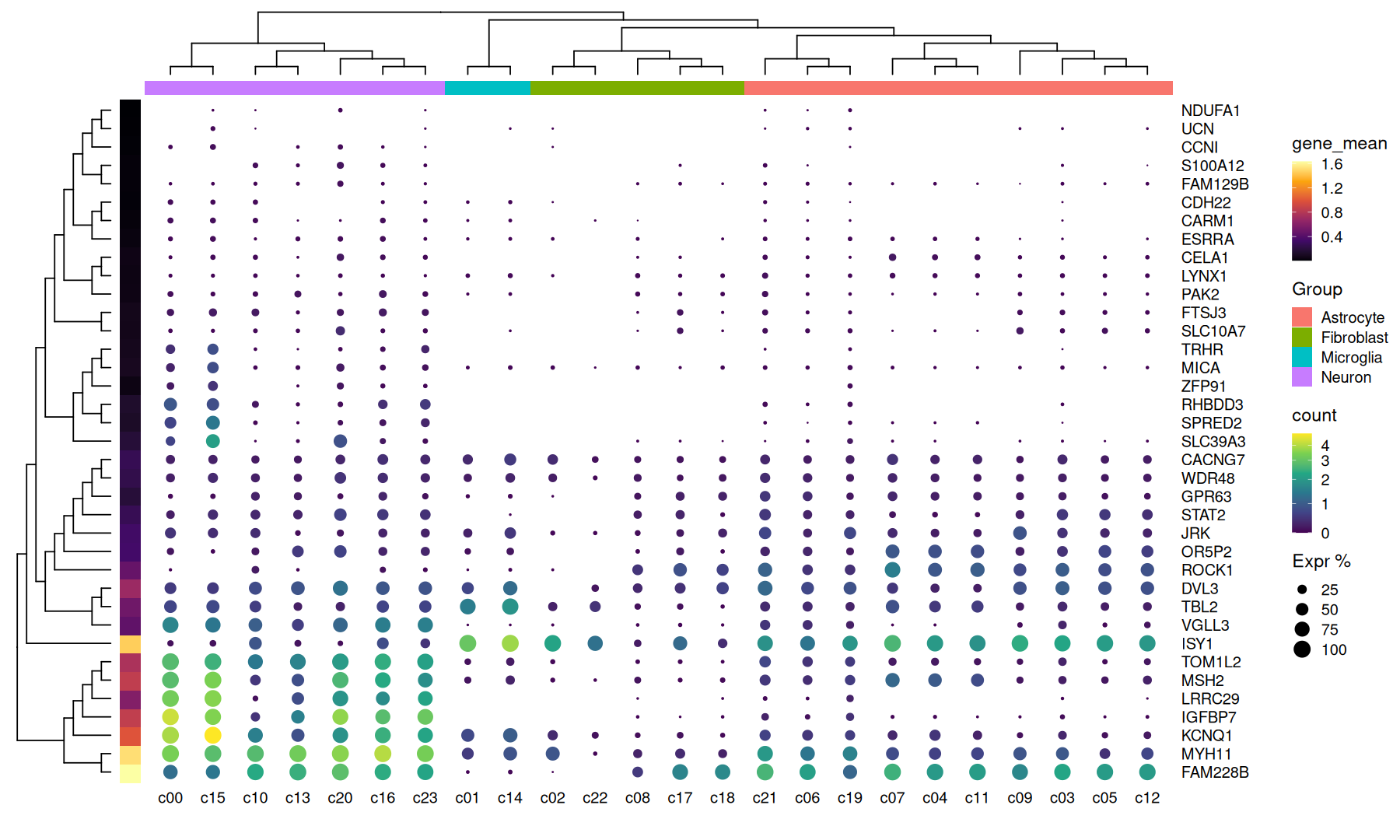
scRNA Heatmap#
data <-
read_tsv(system.file("extdata", "scRNA_dotplot_data.tsv.gz", package="aplot"), col_types='') |>
mutate(expr_perc = 100 * cell_exp_ct/cell_ct) |>
mutate(gene_mean=mean(count), .by=Gene)
head(data,3)
| Gene | cluster | cell_ct | cell_exp_ct | count | Group | expr_perc | gene_mean |
|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <dbl> | <dbl> | <chr> | <dbl> | <dbl> |
| MICA | c00 | 4178 | 910 | 0.2890587840294733 | Neuron | 21.78075634274773 | 0.0840202740097265 |
| MICA | c01 | 3532 | 82 | 0.0232163080407701 | Microglia | 2.32163080407701 | 0.0840202740097265 |
| MICA | c02 | 3320 | 117 | 0.0354171573797353 | Fibroblast | 3.52409638554217 | 0.0840202740097265 |
(
data |>
ggplot(aes(y=Gene, x=cluster, color=count, size=expr_perc)) +
geom_point() +
scale_size(limits=c(0.5, NA), range=c(0,5.5)) +
scale_color_viridis_c(trans='pseudo_log') +
scale_y_discrete(position = 'right') +
theme(axis.ticks = element_blank(),axis.line = element_blank()) +
labs(size='Expr %', x='', y='')
) |>
insert_annot(side='top', var=Group) |>
insert_annot(side='left', var=gene_mean, ggextra=scale_fill_viridis_c(option = 'inferno')) |>
insert_hclust(cluster_by=count, hclust_method='ward.D2')
alternative: ggh4x#
see: https://teunbrand.github.io/ggh4x/articles/PositionGuides.html#dendrograms
library(ggh4x)
clst1 <-
scale(mtcars) |>
dist() |>
hclust("ward.D2")
clst2 <-
scale(mtcars) |>
t() |>
dist() |>
hclust("ward.D2")
df <-
scale(mtcars) |>
as_tibble(rownames = 'model') |>
pivot_longer(names_to = 'vars', values_to = 'value', -model)
ggplot(df, aes(vars, model, fill = value)) +
geom_raster() +
scale_fill_viridis_c() +
coord_cartesian(expand=0) +
labs(y=NULL,x=NULL) +
scale_y_dendrogram(hclust = clst1, guide = guide_dendro(label = FALSE)) +
scale_x_dendrogram(hclust = clst2, position = 'top', guide = guide_dendro(label = FALSE)) +
guides(y.sec = guide_axis(), x.sec=guide_axis())