suppressPackageStartupMessages({
library(tidyverse)
library(cowplot)
library(patchwork)
library(colors3d)
library(prismatic)
library(pals)
theme_set(theme_cowplot())
})
options(repr.plot.width=16,repr.plot.height=10)
Bivariate Color Palette#
code#
map_to_color2d <- function(data, xvar, yvar, colors=c("yellow", "green", "blue", "magenta"), xtrans="rank", ytrans="rank", size=5, na.color='white') {
data |>
summarize(
min_x = min({{xvar}},na.rm=T), max_x = max({{xvar}},na.rm=T),
min_y = min({{yvar}},na.rm=T), max_y = max({{yvar}},na.rm=T)
) |>
with(expand_grid(
x = seq(min_x, max_x, length.out = size),
y = seq(min_y, max_y, length.out = size)
)) |>
mutate(
color = colors2d(tibble(x,y), xtrans = xtrans, ytrans = ytrans, colors=colors)
) |>
ggplot(aes(x,y,fill=color)) +
geom_raster() +
scale_fill_identity() +
labs(x=as_label(enquo(xvar)), y=as_label(enquo(yvar))) +
coord_cartesian(expand=0) -> lgd
data2 <-
select(data, {{xvar}}, {{yvar}}) |>
colors2d(xtrans = xtrans, ytrans = ytrans, colors=colors) |>
bind_cols(data, color=_) |>
mutate(color=ifelse(is.na({{xvar}}) | is.na({{yvar}}), na.color, color))
list(data=data2, legend=lgd)
}
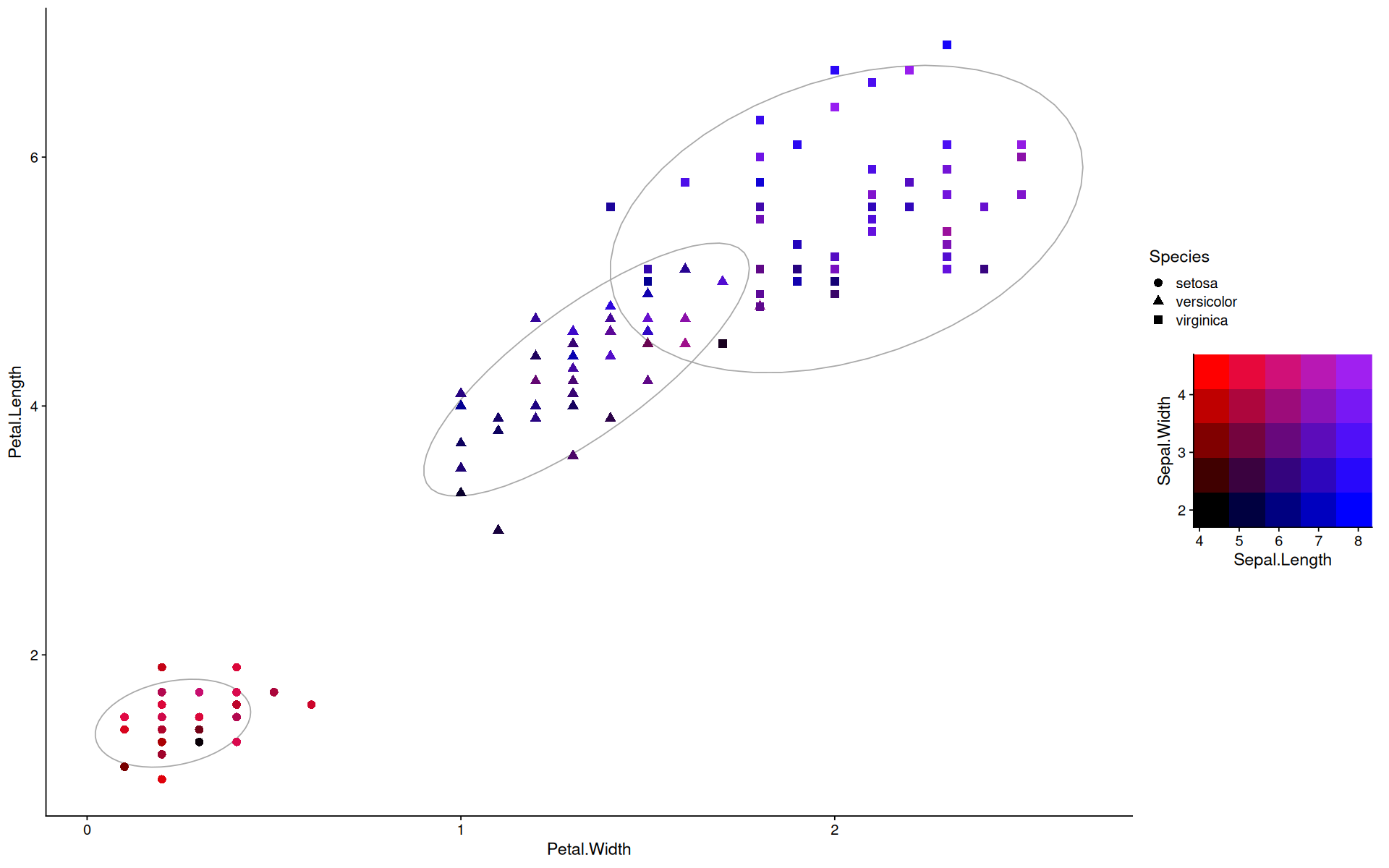
example#
head(iris,3)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species | |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <fct> | |
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
colors <- c('purple', 'blue', 'black', 'red')
r <- map_to_color2d(iris, xvar=Sepal.Length, yvar=Sepal.Width, colors=colors)
ggplot(r$data, aes(y=Petal.Length, x=Petal.Width, color=color, shape=Species, group=Species)) +
stat_ellipse(color='darkgray') +
geom_point(size=3) +
scale_color_identity() +
guides(custom = guide_custom(as_grob(r$legend), width=unit(0.4,'npc'), height=unit(0.4,'npc')))
other palettes#
color(arc.bluepink())
<colors>
#FFFFFFFF #FFE6FEFF #FFBDFFFF #FF80FEFF #E7FFFFFF #D7DAFDFF #D8A6FFFF #C065FEFF #C0FCFDFF #A7CAFFFF #8D7EFDFF #7F65FEFF #74FEFFFF #64C0FFFF #5873FEFF #4B4CFFFF
color(arc.bluepink()[c(16,4,1,13)])
<colors>
#4B4CFFFF #FF80FEFF #FFFFFFFF #74FEFFFF
r <- map_to_color2d(iris, xvar=Sepal.Length, yvar=Sepal.Width, colors=arc.bluepink()[c(16,4,1,13)], size=4)
ggplot(r$data, aes(y=Petal.Length, x=Petal.Width, color=color, shape=Species, group=Species)) +
stat_ellipse(color='darkgray') +
geom_point(size=3) +
scale_color_identity() +
guides(custom = guide_custom(as_grob(r$legend), width=unit(0.4,'npc'), height=unit(0.4,'npc')))
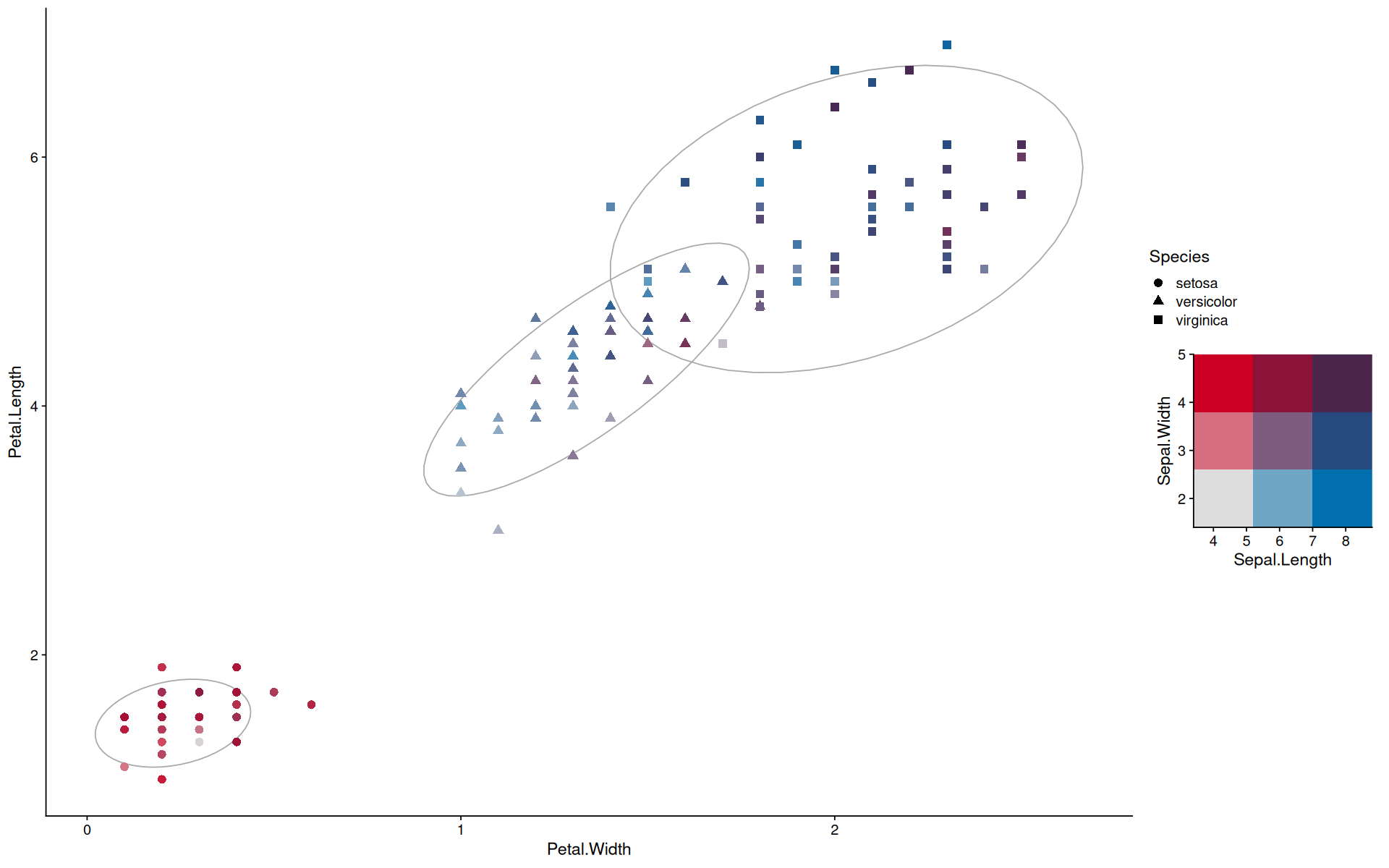
color(tolochko.redblue())
<colors>
#DDDDDDFF #7BB3D1FF #016EAEFF #DD7C8AFF #8D6C8FFF #4A4779FF #CC0024FF #8A274AFF #4B264DFF
color(tolochko.redblue()[c(9,3,1,7)])
<colors>
#4B264DFF #016EAEFF #DDDDDDFF #CC0024FF
r <- map_to_color2d(iris, xvar=Sepal.Length, yvar=Sepal.Width, colors=tolochko.redblue()[c(9,3,1,7)], size=3)
ggplot(r$data, aes(y=Petal.Length, x=Petal.Width, color=color, shape=Species, group=Species)) +
stat_ellipse(color='darkgray') +
geom_point(size=3) +
scale_color_identity() +
guides(custom = guide_custom(as_grob(r$legend), width=unit(0.4,'npc'), height=unit(0.4,'npc')))
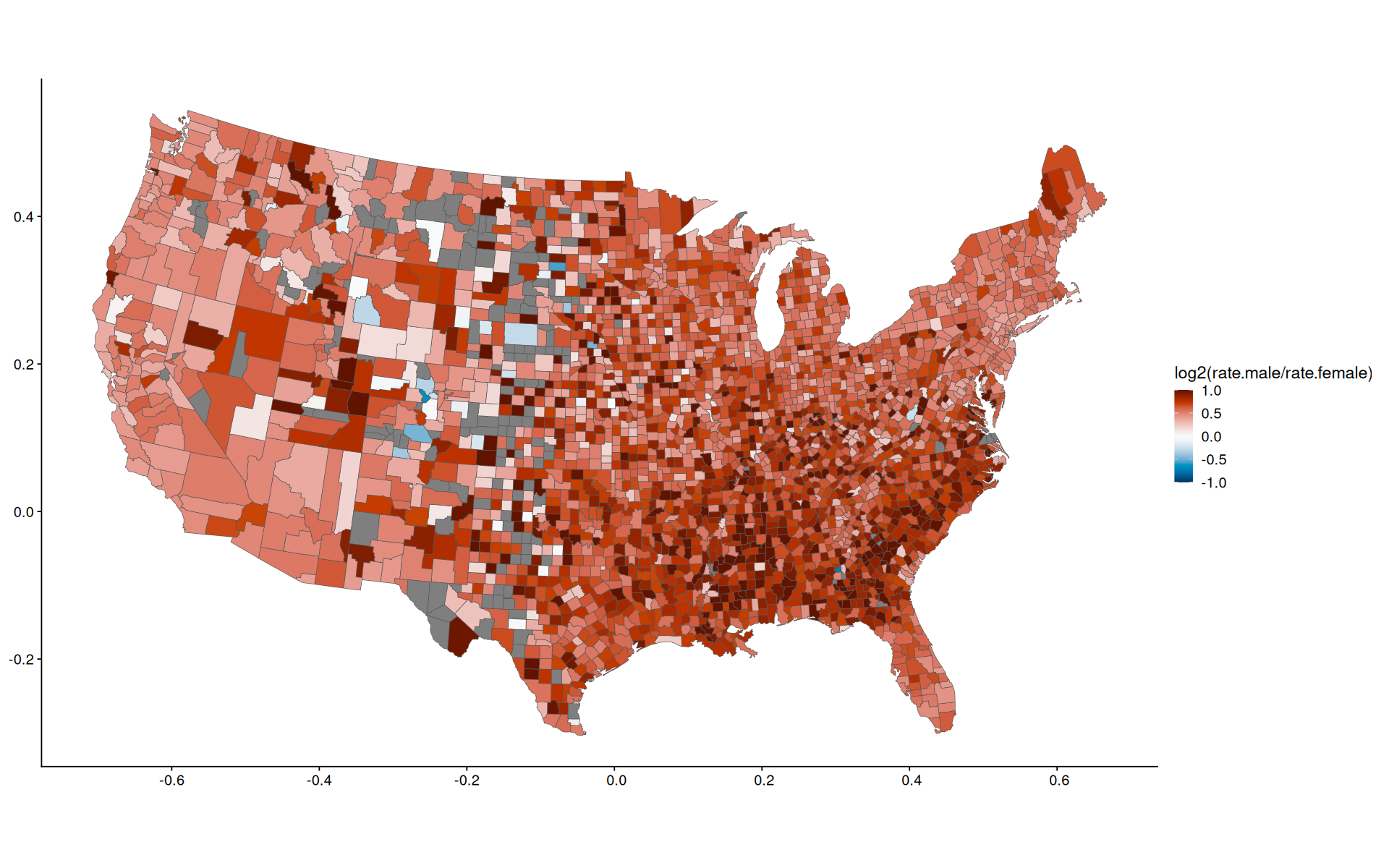
Choropleth#
see also: https://cran.r-project.org/web/packages/pals/vignettes/bivariate_choropleths.html
library(sf)
Linking to GEOS 3.12.2, GDAL 3.9.0, PROJ 9.4.1; sf_use_s2() is TRUE
data(USCancerRates, package='latticeExtra')
data <-
maps::map("county", fill=TRUE, plot =FALSE, projection = "tetra") |>
st_as_sf() |>
as_tibble() |>
left_join(rownames_to_column(USCancerRates, 'ID'), by='ID')
head(data, 3)
| ID | geom | rate.male | LCL95.male | UCL95.male | rate.female | LCL95.female | UCL95.female | state | county |
|---|---|---|---|---|---|---|---|---|---|
| <chr> | <MULTIPOLYGON [°]> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <fct> | <I<chr>> |
| alabama,autauga | MULTIPOLYGON (((0.256464688... | 283.1 | 242.6 | 329.8 | 173.6 | 149.8 | 200.3 | Alabama | Autauga County |
| alabama,baldwin | MULTIPOLYGON (((0.220116483... | 239.2 | 223.5 | 255.8 | 162.0 | 150.7 | 174.1 | Alabama | Baldwin County |
| alabama,barbour | MULTIPOLYGON (((0.288449217... | 335.9 | 288.9 | 389.1 | 185.3 | 157.2 | 217.5 | Alabama | Barbour County |
library(paletteer)
data |>
ggplot(aes(fill=log2(rate.male/rate.female), geometry=geom)) +
geom_sf() +
scale_fill_paletteer_c(palette = 'grDevices::RdBu',limits=c(-1,1), oob=scales::squish, direction = -1)
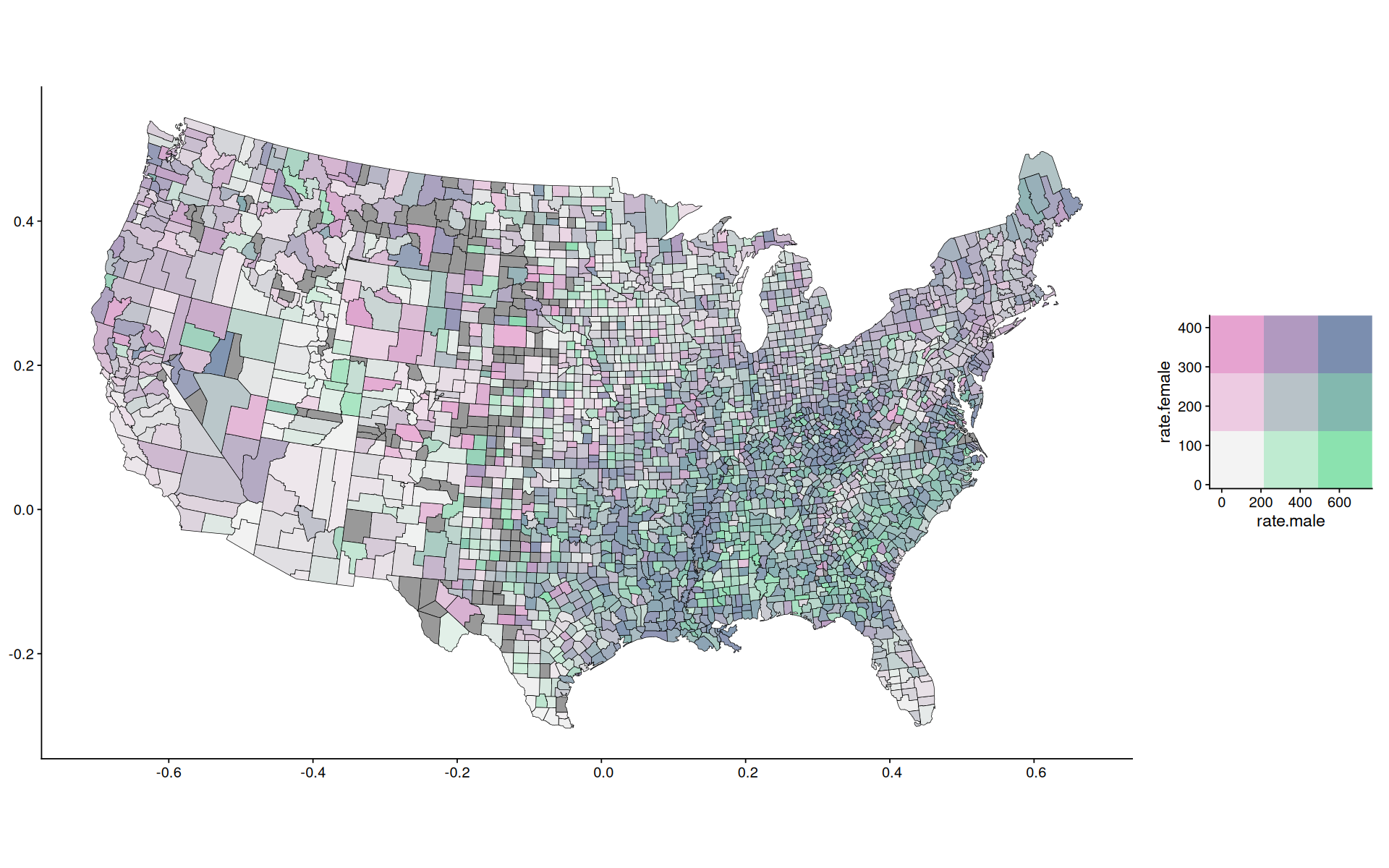
color(stevens.pinkgreen())
<colors>
#F3F3F3FF #C2F1CEFF #8BE2AFFF #EAC5DDFF #9EC6D3FF #7FC6B1FF #E6A3D0FF #BC9FCEFF #7B8EAFFF
color(stevens.pinkgreen()[c(9,3,1,7)])
<colors>
#7B8EAFFF #8BE2AFFF #F3F3F3FF #E6A3D0FF
r <- map_to_color2d(data, xvar=rate.male, yvar=rate.female, colors=stevens.pinkgreen()[c(9,3,1,7)], size=3, na.color='gray60')