suppressPackageStartupMessages({
library(tidyverse)
library(cowplot)
library(broom)
library(dbscan)
theme_set(theme_cowplot())
})
options(repr.plot.width=15,repr.plot.height=9)
Clustering#
k-means clustering#
data("penguins", package = "modeldata")
head(penguins,3)
data <- na.omit(penguins)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex |
|---|---|---|---|---|---|---|
| <fct> | <fct> | <dbl> | <dbl> | <int> | <int> | <fct> |
| Adelie | Torgersen | 39.1 | 18.7 | 181 | 3750 | male |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | 3800 | female |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | 3250 | female |
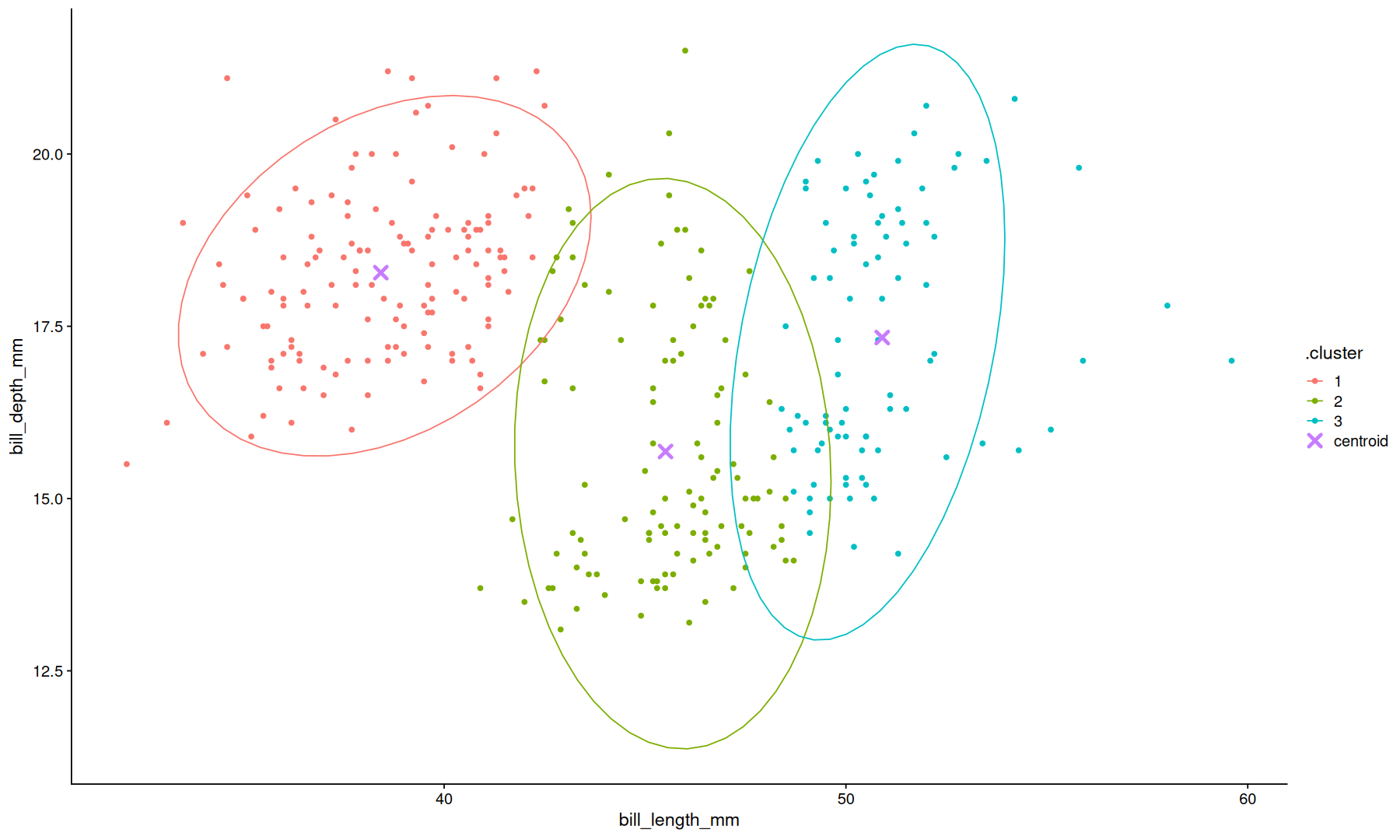
kmeans.obj <-
select(data, bill_length_mm, bill_depth_mm) |>
kmeans(centers=3)
glance(kmeans.obj)
tidy(kmeans.obj)
| totss | tot.withinss | betweenss | iter |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <int> |
| 11216.3616216216 | 2265.06659663866 | 8951.29502498297 | 3 |
| bill_length_mm | bill_depth_mm | size | withinss | cluster |
|---|---|---|---|---|
| <dbl> | <dbl> | <int> | <dbl> | <fct> |
| 38.4242647058823 | 18.2779411764706 | 136 | 904.983750000000 | 1 |
| 45.5098214285714 | 15.6830357142857 | 112 | 742.096964285715 | 2 |
| 50.9035294117647 | 17.3364705882353 | 85 | 617.985882352941 | 3 |
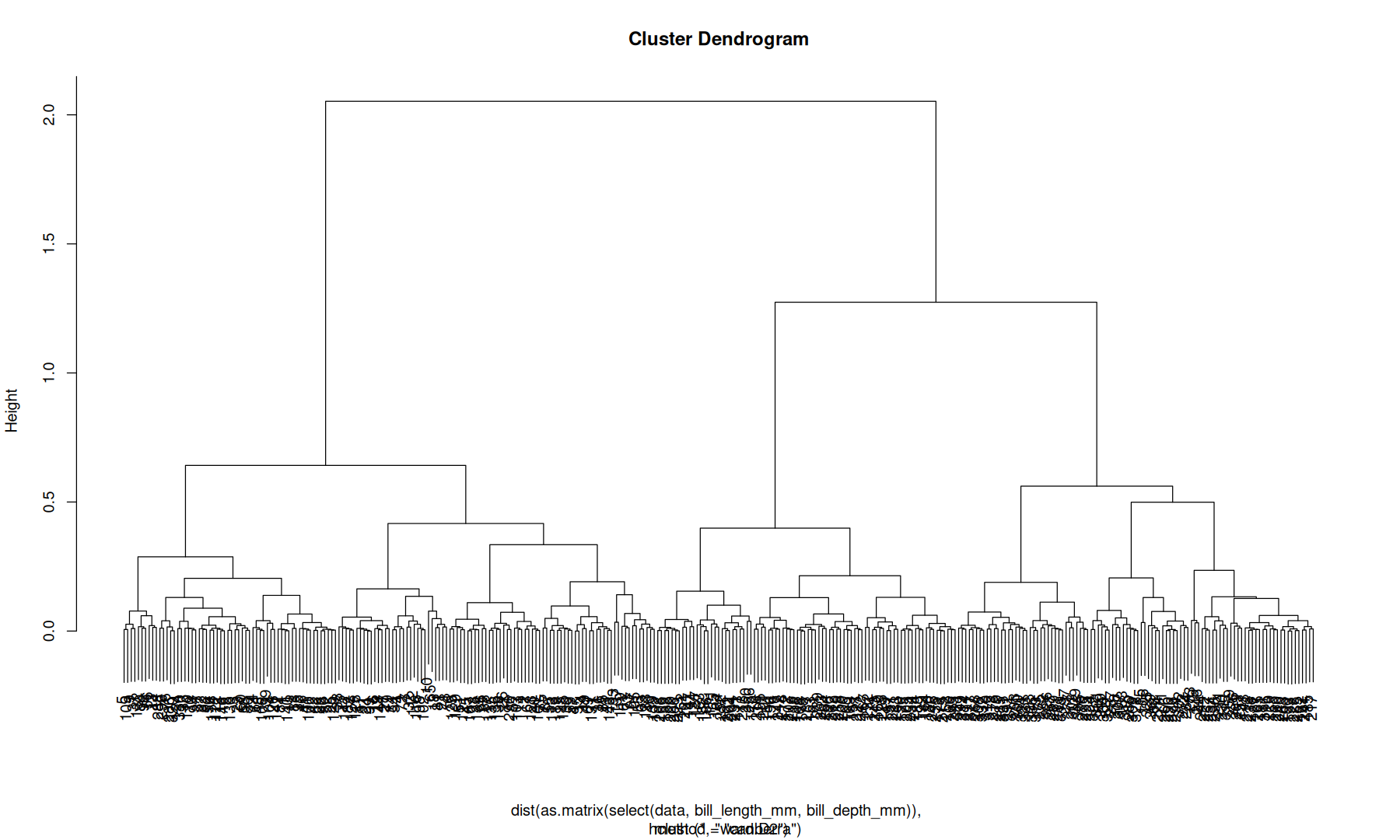
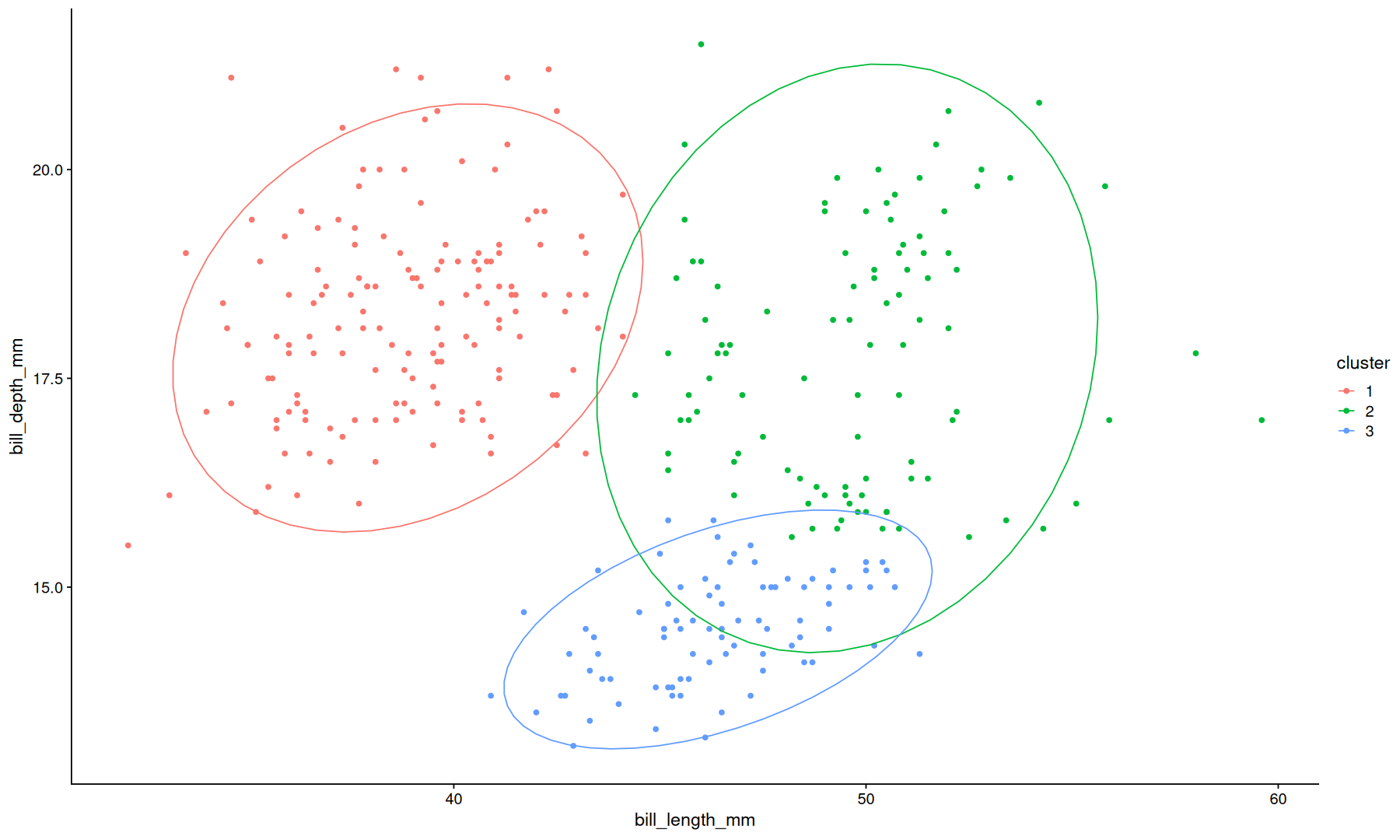
hierarchical clustering#
select(data, bill_length_mm, bill_depth_mm) |>
as.matrix() |>
dist(method = 'canberra') |>
hclust(method='ward.D2') -> hc
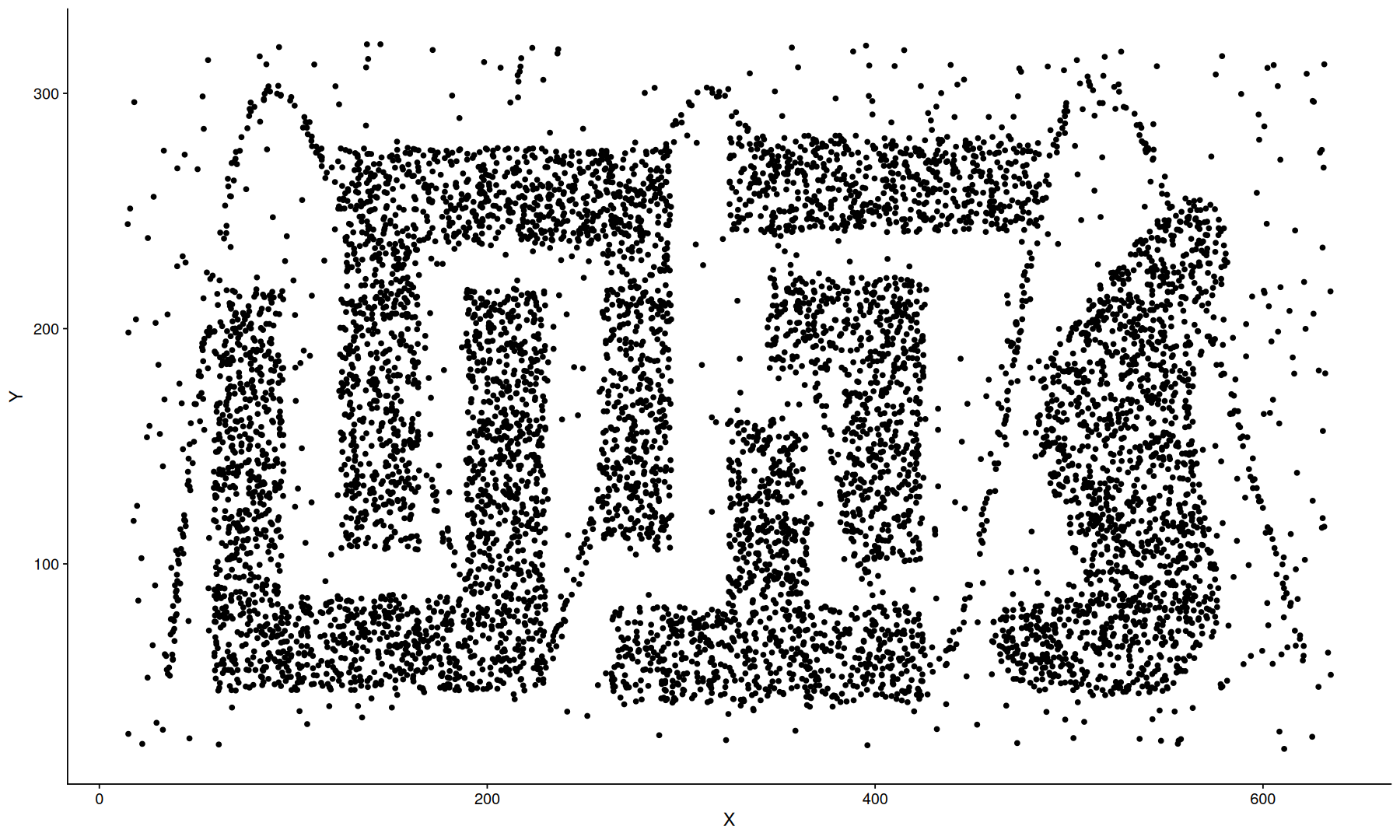
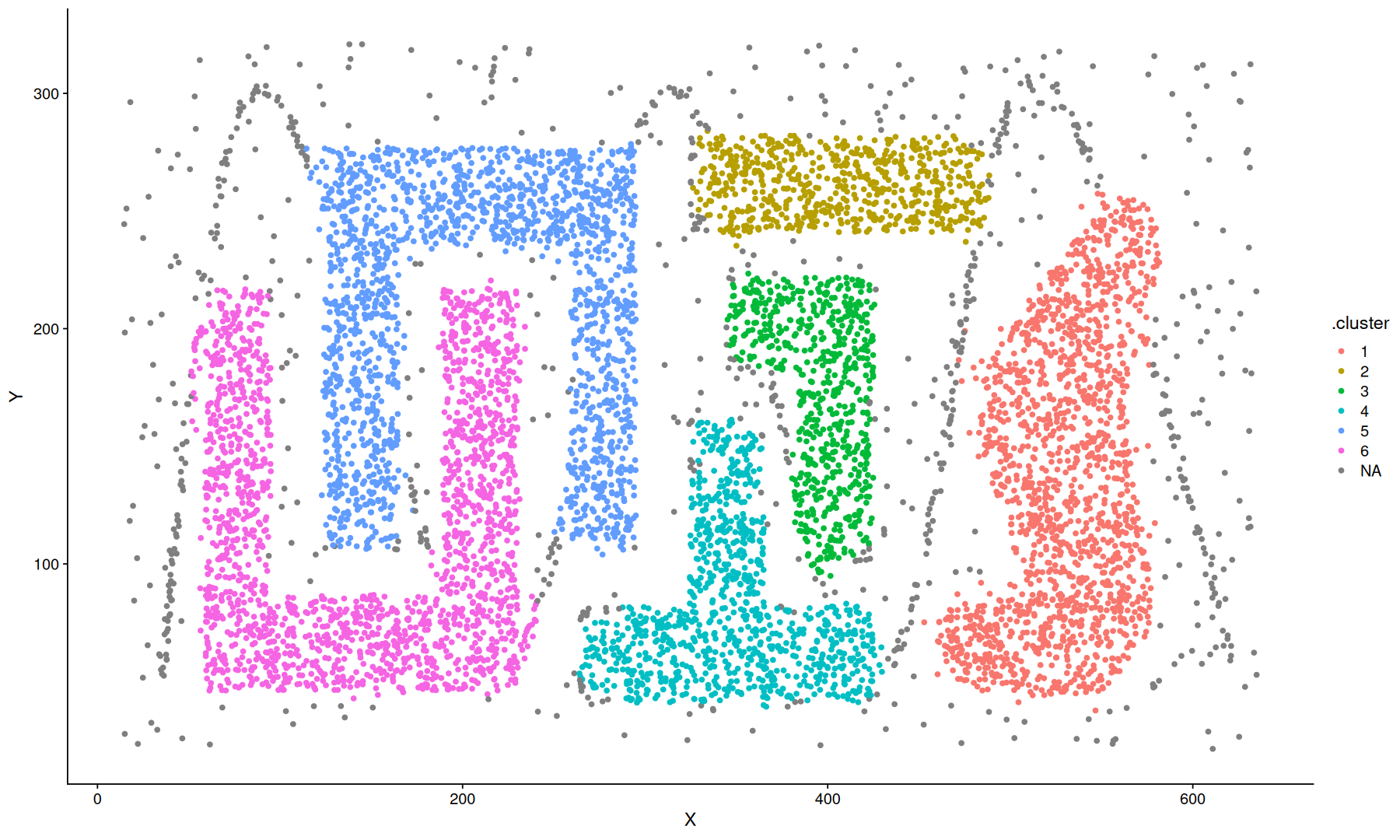
density clustering#
data(DS3, package='dbscan')
dbscan.obj <- hdbscan(DS3, minPts = 25)